WorkShop Avogadro BuildQSAR ChemSketch Dragon5: Difference between revisions
(Imported from text file) |
(revising page for consistent formatting) |
||
| Line 1: | Line 1: | ||
__TOC__ | __TOC__ | ||
'''NDSU-KU Symposium QSAR Workshop''' | '''NDSU-KU Symposium QSAR Workshop''' | ||
| Line 18: | Line 9: | ||
[[File:WorkShop_Avogadro_BuildQSAR_ChemSketch_Dragon5__HTML_ce016f9083bf0bf2.png|308x329px]] | [[File:WorkShop_Avogadro_BuildQSAR_ChemSketch_Dragon5__HTML_ce016f9083bf0bf2.png|308x329px]] | ||
''' | ====== '''1.''' '''ChemSketch''': (Molecule Generation) ====== | ||
# Open ChemSketch and click out of the little popup windows by clicking the X on the top right of the small window. | # Open ChemSketch and click out of the little popup windows by clicking the X on the top right of the small window. | ||
# Next, go to “Tools” and then “Generate.” Select “Structure from SMILES.” | # Next, go to “Tools” and then “Generate.” Select “Structure from SMILES.” | ||
| Line 37: | Line 21: | ||
# Exit ChemSketch. | # Exit ChemSketch. | ||
'''2. Avogadro''': (Molecule Optimization) | ====== '''2. Avogadro''': (Molecule Optimization) ====== | ||
# Open the program Avogadro. | # Open the program Avogadro. | ||
# Go to “File” and “Open” to bring a molecule you created in ChemSketch to Avogadro. Find your molecule that you want to bring to Avogadro and select it to open it. | # Go to “File” and “Open” to bring a molecule you created in ChemSketch to Avogadro. Find your molecule that you want to bring to Avogadro and select it to open it. | ||
| Line 47: | Line 30: | ||
# Exit Avogadro. | # Exit Avogadro. | ||
'''3. Dragon5''': (Descriptor Generation & Data Setup) | ====== '''3. Dragon5''': (Descriptor Generation & Data Setup) ====== | ||
# Open “Dragon5.exe,” exit out of the small windows that pup up. | # Open “Dragon5.exe,” exit out of the small windows that pup up. | ||
# Select “Calculate Descriptors” and then select all the “.mol2” files that you want to calculate. Use the ‘Ctrl’ button when selecting the files so all desired files can be selected. | # Select “Calculate Descriptors” and then select all the “.mol2” files that you want to calculate. Use the ‘Ctrl’ button when selecting the files so all desired files can be selected. | ||
| Line 63: | Line 45: | ||
# Leave the DESCRIPTOR.txt file open | # Leave the DESCRIPTOR.txt file open | ||
'''4. BuildQSAR''': (Model Development) | ====== '''4. BuildQSAR''': (Model Development) ====== | ||
<ol> | <ol> | ||
<li><p>Open “BuildQSAR.”</p></li> | <li><p>Open “BuildQSAR.”</p></li> | ||
| Line 88: | Line 69: | ||
<li><p>That is your developed models.</p></li></ol> | <li><p>That is your developed models.</p></li></ol> | ||
=== '''Descriptor Information''': === | |||
# So each model (AKA equation) has a certain number of descriptors in it. In this example there are 3. | |||
<math display="block">Y = -1.6198 (\pm 1.1586) X_{269} + 0.0110 (\pm 0.0016) X_{631} - 0.0336 (\pm 0.0094) X_{634} + 0.8483 (\pm 0.3386})</math> | |||
<li>To find the meaning of these descriptors go to your Excel file and above your descriptor data create a row that is listed from X1 to X####. X1 is above the first descriptor, usually MW. The #’s are the number of descriptors in one row. | |||
</li> | |||
< | <li>On Excel find the descriptors that are listed in the model. Find the abbreviation under the X value. Example X269 corresponded to the abbreviation “MATS3e” | ||
< | </li> | ||
<li>Once you have all the descriptor abbreviations then go back to Dragon. | |||
</li> | |||
< | <li>Select “ Descriptor Search” | ||
< | </li> | ||
<li>Type an abbreviation you found from the Excel file. | |||
</li> | |||
<li>Press “Search” | |||
</li> | |||
< | <li>This will give the descriptor information. | ||
<li> | |||
< | |||
<li>< | |||
</li> | </li> | ||
<li | <li>You can also click on this link to find what the abbreviation means | ||
# <li><u>http://www.talete.mi.it/products/dragon_molecular_descriptor_list.pdf</u> | |||
</li></li> | |||
<li | |||
</li | |||
Revision as of 19:54, 26 September 2022
NDSU-KU Symposium QSAR Workshop
Purpose: Using ChemSketch, Avogadro, Dragon5, and Build QSAR programs to generate structures, descriptors, and models.
1. ChemSketch: (Molecule Generation)
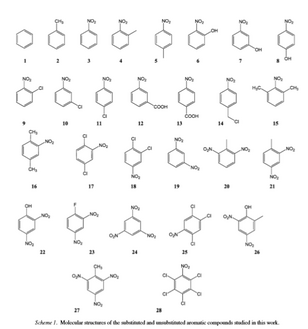
- Open ChemSketch and click out of the little popup windows by clicking the X on the top right of the small window.
- Next, go to “Tools” and then “Generate.” Select “Structure from SMILES.”
- Type in the SMILES Notation from above.
- Push “OK.”
- Left click where you want to make the molecule.
- Once the desired molecule is drawn then go to “Tools” and click “Clean Structure.” This will make it easier for the other programs to read what you created.
- Then go to “File” then “Save as” and choose the “KU WorkShop Practice” Folder on the DESKTOP.
- Save ChemSketch structure as “#.mol” where # is the number associated with the table above.
- Repeat steps to create the desired number of structures. Save them as individual *.mol Files.
- Exit ChemSketch.
2. Avogadro: (Molecule Optimization)
- Open the program Avogadro.
- Go to “File” and “Open” to bring a molecule you created in ChemSketch to Avogadro. Find your molecule that you want to bring to Avogadro and select it to open it.
- A small window will pop up about 3D coordinates and a rough sketch. Click “Yes.”
- Once the molecule is on Avogadro then go to “Extensions” and select “Optimize geometry.”
- Then go to “File” then “Save as” and choose the “KU WorkShop Practice” Folder on the DESKTOP.
- Save the files as *.mol2 for all the individual molecules you created.
- Exit Avogadro.
3. Dragon5: (Descriptor Generation & Data Setup)
- Open “Dragon5.exe,” exit out of the small windows that pup up.
- Select “Calculate Descriptors” and then select all the “.mol2” files that you want to calculate. Use the ‘Ctrl’ button when selecting the files so all desired files can be selected.
- Press the green check mark “OK” when all desired files are selected.
- Choose the desired descriptors you want calculated for these molecule files. “X” means checked. Then press RUN.
- Press Continue when the small window pops up.
- A yellow window pops up and gives information about the calculations. If no errors are listed on the yellow window then exit out of it.
- Select “Save Descriptors.”
- Make sure “Constant Variables” & “Near-Constant Variables” are selected “x.” Select “Pair Correlation” and pick .95 or something around that number.
- Press “Save” and save as “DESCRIPTORS.txt” file.
- Exit Dragon
- Find the “DESCRIPTOR.txt” file you saved and open with Notepad++. You can do this by finding the file and right clicking on the file. Select “Edit with Notepad++” to open it in Notepad++.
- ***VERY IMPORTANT Write the number of descriptors, the number is listed in 2nd Row, 3rd Column.
- Leave the DESCRIPTOR.txt file open
4. BuildQSAR: (Model Development)
Open “BuildQSAR.”
Go to “File” then “New.”
Add a Title to “Dataset Title.”
Change the number of “Compounds” to the number of molecules you have created AND ADD 4. For example, if you have 10 molecules put in the number 14. Change the number of “Descriptors” to the number you got from the *.txt file AND ADD 2. So if you have 632 descriptors then type 634.
Click “Ok.”
From your DESCRIPTOR.txt file select all the information by pressing “Ctrl + A.” Copy the selected information by pressing “Ctrl + C”
In buildQSAR select the top left BLUE square and paste the data by “right clicking” using the mouse and select “PASTE”
Delete the columns and rows that have words and incomplete information. Usually that is the first 4 rows. You can do this by left clicking One data point from those columns or rows then select “Dataset”, “Remove.” The Row is removed if you press “compound.” The Column is removed if you press “descriptor”.
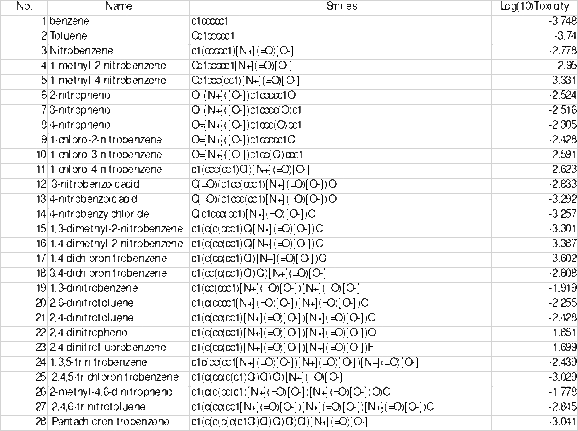
In the yellow (Y1) column input the data from the Table above which shows the Log(10)Toxicity
Go to “QSAR” then “Variable Selection” then “Systematic Search” or “Genetic Algorithm.” (note: Choose Genetic Algorithm only when you need 4, 5 or higher number of variables in the model).
A small popup window will pop up. Make sure the 2 boxes under “Cross Validation” are checked.
The correlation criteria can change but if uncertain on a number then put 0.6 as default.
For “Genetic Algorithm:”
“Descriptors per Model,” this is usually calculated using the 5-1 rule. The 5-1 rule relates the number of molecules you have to the number “Variables AKA Descriptors” in your “Model oKA Equation.” Example: 5-1 rule is used on 24 molecules you should have 4 in the “Descriptors per model” section. ** DON’T ROUND UP **
“No. of generations” can vary 200-500), but 200 is an okay to use as a default number.
“Models per Generation” should be at least 3 (better to have between 5-10).
Press “Run.”
When complete “Double Click” on any of the cells in the first row.
That is your developed models.
Descriptor Information:
- So each model (AKA equation) has a certain number of descriptors in it. In this example there are 3.
Failed to parse (syntax error): {\displaystyle Y = -1.6198 (\pm 1.1586) X_{269} + 0.0110 (\pm 0.0016) X_{631} - 0.0336 (\pm 0.0094) X_{634} + 0.8483 (\pm 0.3386})}