Generating Descriptors Workflow: Difference between revisions
(Imported from text file) |
m (Sysadmin moved page GeneratingDescriptors Model ChemSketch HyperChem OpenBabel Avogadro Dragon BuildQSAR to Generating Descriptors Workflow without leaving a redirect: Page rename) |
(No difference)
| |
Revision as of 21:03, 26 September 2022
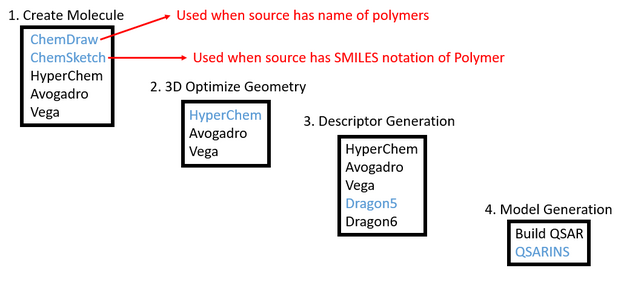
Procedure Document for using Chem Sketch, Avogadro, Dragon, Excel, Hyperchem, & Build QSAR to obtain Descriptors.
Purpose: Using ChemSketch, ChemDraw, HyperChem, Avogadro, Dragon5, Build QSAR, and QSARINS programs to generate structures, descriptors, and models. Also, OpenBabel is used for converting files from one type to another.
Chem Sketch: Special can convert SMILES to structure and structure to SMILES.
- Open Chem Sketch and click out of the little popup windows by clicking the X on the top right of the small window.
- Draw the desired molecule in skeletal format.
- Once the desired molecule is drawn then go to “Tools” and click “Clean Structure.” This will make it easier for the other programs to read what you created.
- Next, go to “Tools” again and then “Generate.” You will have to generate the “Name of Structure,” “Smiles Notation,” and “InChi for Structure.”
- Open a blank document on Excel. For the molecule created copy and paste the three things you generated. These three things will help keep your data organized. You may not need to refer to all three of these again but it is best to have them ready on hand as a precaution. You will use the “Name of Structure” for naming the files. ** Make sure you assign a number to each molecule. If you have 10 molecules just assign them numbers 1-10. Once again this will help with organizing your data. Organization is a big thing in this work!
- After the three things you generated are on Excel and you assigned the molecule a number. Then delete the names off of chem sketch. Keep the structure and save it as an “MDL Molfiles (*.mol).” The file name can be whatever but it is preferred to follow this format. “Assigned # Name of Structure” Example… “33 2-ethenyl-5-nitrofuran.mol” The “.mol” should be automatically added once you select “MDL molfiles (*.mol).
- Exit Chem Sketch.
Chem Draw: SPECIAL can convert name of structure to a structure.
- ChemDraw has a special feature that if given a name of a polymer, it will generate the monomer unit for you.
- Open ChemDraw Professional
- Go to “Structure”
- Select “Convert Name to Structure”
- Type in the name of the polymer without “poly” for example. “Polybutane” only type “butane.”
- It will give you a generalized version. This will assist in giving you the correct monomer unit or just generalized version.
- *** ONLY HAVE STURCTURE IN “*.MOL” FILE*** Delete the text after the model is generated because the text will cause issues later
- NEXT STEPS ARE FOR CREATION OF STRUCTURE AND NAME REGULAR WAY LIKE DRAWING
- Draw the desired molecule
- Select the model by pressing the logo near the top left of the screen

- Once selected go to “Structure” and click “Clean up Structure.” This will make it easier for the other programs to read what you created.
- To generate the name of this structure go to “Structure” and press “Convert structure to name.”
- Open a blank document on Excel. For the molecule created copy and paste the name you generated. This will help keep your data organized.
- After the model is generated and the name is on Excel, assign a number to the molecule on excel. Then delete the names off of ChemDraw. Keep the structure and save it as an “MDL Molfiles (*.mol).” The file name can be whatever but it is preferred to follow this format. “Assigned # Name of Structure” Example… “33 2-ethenyl-5-nitrofuran.mol” The “.mol” should be automatically added once you select “MDL molfiles (*.mol).
- *** ONLY HAVE STURCTURE IN “*.MOL” FILE*** Delete the text after the model is generated because the text will cause issues later
- Exit Chem Draw.
Avogadro:
After doing these steps for all the molecules you wanted to create then open the program Avogadro.
Go to “File” and “Open” to bring a molecule you created in ChemSketch to Avogadro. Find your molecule that you want to bring to Avogadro and select it to open it. Click “Yes” when a small window pops up about 3D coordinates and a rough sketch.
Once the molecule is on Avogadro then go to “Extensions” and select “Optimize geometry.”
* There is another way to Optimize Geometry How do we do it again???
Save the file as “Sybyl Mol2 (*.mol2)”
Exit Avogadro.
Dragon5: (IT IS OKAY TO USE *.mol and *.hin or any other format usable on Dragon, no issues what so ever)
Open “Dragon5.exe,” exit out of the small windows that pup up.
Select “Calculate Descriptors” and then select all the “.mol2” files that you want to calculate. Use the ‘Ctrl’ button when selecting the files so all desired files can be selected.
Press the green check mark “OK” when all desired files are selected.
Choose the desired descriptors you want calculated for these molecule files. “X” means checked. Then press RUN.
Press Continue when the small window pops up.
A yellow window pops up and gives information about the calculations. If no errors are listed on the yellow window then exit out of it.
Select “Save Descriptors.”
Make sure “Constant Variables” & “Near-Constant Variables” are selected “x.” Select “Pair Correlation” and pick .95 or something around that number.
Press “Save” and save as “.txt” file.
Leave Dragon open for future use.
Find the file you saved and open with Notepad++. You can do this by finding the file and right clicking on the file. Select “Edit with Notepad++” to open it in Notepad++.
In Notepad++, select all the information and copy onto an EXCEL worksheet. If desired you can delete the first 2 rows of the information on Notepad++. It is recommended to write the number of descriptors. This will help you later. It is listed in the 2nd row, 3rd column.
Once all the information is saved onto Excel then exit out of the Notepad++ file.
Splitting to Training and Test sets
In Excel worksheet, split the whole set to training and test sets by sorting your molecules according to “Experimental values” or “Y” column, in descending or ascending order. After sorting, select every 5th (20% of the set) or every 4th (25% of the set) compound (row) and then copy it to another excel sheet, naming it as a Test set. Remove the test compounds from the initial set to make a training set.
Use the obtained training set to build a dataset in BuildQSAR.
BuildQSAR:
Open “BuildQSAR.”
Go to “File” then “New.”
Add a Title to “Dataset Title.”
Change the number of “Compounds” to the amount in your data. Change the number of “Descriptors” you have. The number of descriptors was obtained from the Notepad++ information.
Click “Ok.”
Input observed data into the yellow column. The yellow column is for the observed information from other sources such as research papers.
** The observed data should be put into Excel at this point.
Copy and paste the descriptor information from Excel into the blue cells of BuildQSAR.
Go to “QSAR” then “Variable Selection” then “Systematic Search” or “Genetic Algorithm.” (note: Choose Genetic Algorithm only when you need 4, 5 or higher number of variables in the model).
A small popup window will pop up. Make sure the 2 boxes under “Cross Validation” are checked. The correlation criteria can change but if uncertain on a number then put 0.6 as default.
“Descriptors per Model,” this is usually calculated using the 5-1 rule. The 5-1 rule relates the number of molecules you have to the number “Variables AKA Descriptors” in your “Model oKA Equation.” Example: 5-1 rule is used on 24 molecules you should have 4 in the “Descriptors per model” section. ** DON’T ROUND UP **
“No. of generations” can vary 200-500), but 200 is an okay default number to have.
“Models per Generation” should be at least 3 (better to have between 5-10).
Press “Run.”
Double Click on any of the cells in the first row.
A pop up window with a “Model aka equation” will pop up.
Copy and paste the model and information in the “()” onto Excel.
Close out of the window with the model information.
Copy the model and () information from all three rows.
These rows are the different models the BuildQSAR generated for you.
Descriptor Information:
So each model (AKA equation) has a certain number of descriptors in it. In this example there are 3.
Y1 = - 1.6198 (± 1.1586) X269 + 0.0110 (± 0.0016) X631 - 0.0336 (± 0.0094) X634 + 0.8483 (± 0.3386)
To find the meaning of these descriptors go to your Excel file and above your descriptor data create a row that is listed from X1 to X####. X1 is above the first descriptor, usually MW. The #’s are the number of descriptors in one row.
On Excel find the descriptors that are listed in the model. Find the abbreviation under the X value. Example X269 corresponded to the abbreviation “MATS3e”
Once you have all the descriptor abbreviations then go back to Dragon.
Select “ Descriptor Search”
Type an abbreviation you found from the Excel file.
Press “Search”
This will give the descriptor information.
Test Set predictions
Use obtained model to calculate “Predicted Experimental values” for the Test set.
For that, find the descriptors selected by the model, find them among test set descriptors, and then calculate “Predicted Experimental values” by applying mathematical model from training set and values of selected descriptor for each compound in the test set.
Do the same for all models, from 1 to 5 variables. Calculate predictive R2 values by correlation of “Experimental values” and Predicted Experimental values” in Excel (function CORR( )).
Build a graph for all models from 1 to 5 variables, by drawing on axes X No of model and on axes Y the values R2 for each model (separate lines for training and for the tests sets).
Select the best model by finding the maximum value of R2 for the test set models.
Open Babel GUI:
This section depends on conversion of one file type to another if needed. Usually this is done to get HOMO LUMO (Extra descriptors) from Hyperchem (another 3D software) which has file types of “*.hin” Also, this is used to convert file type to smiles notation if needed. In this example we are converting “*.mol” files to Smiles format notation and “*.hin” format. “*.hin” is for HyperChem
Open “Open Babel GUI”
On the top left of the window that opens chose the file type that you have. For example “mol—MDL MOL format is a common file extension.”
Once chosen you can select the way you want the data read in OpenBabel. In this example the “Use this format for all input files (ignore file extensions)” will be checked.
Clicking on the left side “…” box you can choose the file you would like converted.
Once the file was chosen it should be in the bar to the left of the “…” box
Under the very top right comment of “OUTPUT FORMAT” chose what you would like converted to. In this example chose “smiles - - SMILES format”
Check “Add Hydrogens (make explicit) in the middle column
Check “Generate 3D coordinates” in the middle column
Simply press convert and the data you want should be listed in the right column.
Copy the Smiles notation and save it onto another file such as excel or notepad. This is one example of what a smiles notation looks like “OC(F)(F)[C@H](F)C(F)(F)F”
The same step as 62 can be used to create the “*.hin” format notation. This is done by choosing “hin - - HyperChem HIN format” under OUTPUT FORMAT.
Make sure “Output below only (no output file)” and “Display in firefox” are NOT checked. These are on the right side column or AKA output column.
Before pressing Convert you need to specify an output file and location. A good idea is to name it exactly the same as the input fil except change the “.mol” ending file to “.hin” (THE “.hin” NEEDS TO BE INCLUDED IN THE OUTPUT FILE)
Press convert
The output file should be in the location with the name specified.
HyperChem:
This section depends on the need for quantum descriptors such as HOMO, LUMO, and Total energy.
Open “HyperChem”
Open a file that has “.hin” extension
Press “Setup” on the tool bar
Press “Semi-empirical”
Press “RM1”
Press “OK
Press “Compute” on the tool bar
Select “Geometry Optimization”
For regular work make sure “Polak –Ribiere (Conjugate gradiant)” is selected.
Have 0.1 in the top bar of RMS gradiant and in the bottom bar 600.
Have “In vacuo” selected
For the “Screen refresh period” use 1 for cycles.
Steps 78-81 should be default settings
Press “OK”
Once the optimization has stopped (Numbers on the bottom right or left have stopped moving) then select “Compute”
Select “Properties”
All the information in “Properties” can be used as quantum descriptors if needed
By Pressing “Details” you will get more information and these are also used as descriptors if needed.
Exit out of “Properties”
Press “Compute”
Select “Orbitals”
the screen that pops up shows 2-4 different columns, the left being Alpha orbitals and the right being Beta orbitals. The pink/purple column is simply the orbitals above the HOMO-LUMO Gap, and the green is simply below the HOMO LUMO Gap
Each line in these columns gives you the energy in “eV” for that orbital.
If you zoom into the bottom most line of the pink/purple column that is LUMO- Lowest unoccupied molecular orbital. If you select the bottom most line of the pink/purple column, it will give you a value in the “Energy” bar as “eV”. That is LUMO.
If you zoom into the top most line of the green column that is HOMO- Highest Occupied Molecular Orbital. If you select the top most like of the green column it will give you a value in the “Energy” bar as “eV”. That is HOMO
The difference of these energies is the HOMO-LUMO Gap.
If desired, you can simply “Plot” isosurfaces of the orbital that is selected by pressing the “Plot” button on the bottom left of the Orbitals panal.
Press “OK” when done with this screen.
To get further Descriptors
Select “Compute”
Select “ QSAR Properties” at the bottom
In this window you can select any of the properties that you desired to compute by simply selecting the property and press compute. The value will be shown at the bottom of the “QSAR Properties” window.
Scripts are also available to be written for Hyperchem if desired, you can do this via following Lesson 17 or 18 in this manual. http://www.chemistry-software.com/pdf/Hyperchem_full_manual.pdf
If manual doesn’t show up just google “Hyperchem Manual”If It says your Molecule has issues before Geometry optimization such as Valence error go to Select
Select All
Select again
“Add H & Model Build”
This corrects the error
Additionally, If molecules are large and “Semi-Empirical” geometry optimization takes too much time then molecular mechanics is needed
When doing the molecular mechanics it is necessary to make sure the atom “type” is corrected.
To check if atom type is correct go to “Display”
“Labels”
“type”
If you see “**” on the atoms then hyperchem needs to recalculate the atoms to get C, H, O, and other atoms.
Go to “setup”
“Molecular Mechanics”
Select “MM+”
Click on “Options”
Press OK
Click on “Components”
Press OK
Then press “OK” on the same window you selected “MM+”
You should get a popup window saying the atom types are going to be recalculated
Press “OK”
This should change the “**” to letters such as C, H, O
If it doesn’t initially then try going to labels and pressing “none.” Then repeat steps 114 a-b
If that doesn’t work then select the semi-empirical. Once selected redo steps 114a-d
Use RHF not UHF